Segmentation (image processing)
In computer vision, segmentation is the process of partitioning a digital image into multiple segments (sets of pixels, also known as superpixels). The goal of segmentation is to simplify and/or change the representation of an image into something that is more meaningful and easier to analyze.[1] Image segmentation is typically used to locate objects and boundaries (lines, curves, etc.) in images. More precisely, image segmentation is the process of assigning a label to every pixel in an image such that pixels with the same label share certain visual characteristics.
The result of image segmentation is a set of segments that collectively cover the entire image, or a set of contours extracted from the image (see edge detection). Each of the pixels in a region are similar with respect to some characteristic or computed property, such as color, intensity, or texture. Adjacent regions are significantly different with respect to the same characteristic(s).[1] When applied to a stack of images, typical in Medical imaging, the resulting contours after image segmentation can be used to create 3D reconstructions with the help of interpolation algorithms like Marching cubes.
Applications
Some of the practical applications of image segmentation are:
- Medical imaging[2]
- Locate tumors and other pathologies
- Measure tissue volumes
- Computer-guided surgery
- Diagnosis
- Treatment planning
- Study of anatomical structure
- Locate objects in satellite images (roads, forests, etc.)
- Face recognition
- Iris recognition
- Fingerprint recognition
- Traffic control systems
- Brake light detection
- Machine vision
- Agricultural imaging – crop disease detection
Several general-purpose algorithms and techniques have been developed for image segmentation. Since there is no general solution to the image segmentation problem, these techniques often have to be combined with domain knowledge in order to effectively solve an image segmentation problem for a problem domain.
Thresholding
The simplest method of image segmentation is called the thresholding method. This method is based on a clip-level (or a threshold value) to turn a gray-scale image into a binary image.
The key of this method is to select the threshold value (or values when multiple-levels are selected). Several popular methods are used in industry including the maximum entropy method, Otsu's method (maximum variance), and et al. k-means clustering can also be used.
Clustering methods
The K-means algorithm is an iterative technique that is used to partition an image into K clusters. The basic algorithm is:
- Pick K cluster centers, either randomly or based on some heuristic
- Assign each pixel in the image to the cluster that minimizes the distance between the pixel and the cluster center
- Re-compute the cluster centers by averaging all of the pixels in the cluster
- Repeat steps 2 and 3 until convergence is attained (e.g. no pixels change clusters)
In this case, distance is the squared or absolute difference between a pixel and a cluster center. The difference is typically based on pixel color, intensity, texture, and location, or a weighted combination of these factors. K can be selected manually, randomly, or by a heuristic.
This algorithm is guaranteed to converge, but it may not return the optimal solution. The quality of the solution depends on the initial set of clusters and the value of K.
In statistics and machine learning, the k-means algorithm is a clustering algorithm to partition n objects into k clusters, where k < n. It is similar to the expectation-maximization algorithm for mixtures of Gaussians in that they both attempt to find the centers of natural clusters in the data. The model requires that the object attributes correspond to elements of a vector space. The objective it tries to achieve is to minimize total intra-cluster variance, or, the squared error function. The k-means clustering was invented in 1956. The most common form of the algorithm uses an iterative refinement heuristic known as Lloyd's algorithm. Lloyd's algorithm starts by partitioning the input points into k initial sets, either at random or using some heuristic data. It then calculates the mean point, or centroid, of each set. It constructs a new partition by associating each point with the closest centroid. Then the centroids are recalculated for the new clusters, and algorithm repeated by alternate application of these two steps until convergence, which is obtained when the points no longer switch clusters (or alternatively centroids are no longer changed). Lloyd's algorithm and k-means are often used synonymously, but in reality Lloyd's algorithm is a heuristic for solving the k-means problem, as with certain combinations of starting points and centroids, Lloyd's algorithm can in fact converge to the wrong answer. Other variations exist, but Lloyd's algorithm has remained popular, because it converges extremely quickly in practice. In terms of performance the algorithm is not guaranteed to return a global optimum. The quality of the final solution depends largely on the initial set of clusters, and may, in practice, be much poorer than the global optimum. Since the algorithm is extremely fast, a common method is to run the algorithm several times and return the best clustering found. A drawback of the k-means algorithm is that the number of clusters k is an input parameter. An inappropriate choice of k may yield poor results. The algorithm also assumes that the variance is an appropriate measure of cluster scatter.
Compression-based methods
Compression based methods postulate that the optimal segmentation is the one that minimizes, over all possible segmentations, the coding length of the data.[3][4] The connection between these two concepts is that segmentation tries to find patterns in an image and any regularity in the image can be used to compress it. The method describes each segment by its texture and boundary shape. Each of these components is modeled by a probability distribution function and its coding length is computed as follows:
- The boundary encoding leverages the fact that regions in natural images tend to have a smooth contour. This prior is used by huffman coding to encode the difference chain code of the contours in an image. Thus, the smoother a boundary is, the shorter coding length it attains.
- Texture is encoded by lossy compression in a way similar to minimum description length (MDL) principle, but here the length of the data given the model is approximated by the number of samples times the entropy of the model. The texture in each region is modeled by a multivariate normal distribution whose entropy has closed form expression. An interesting property of this model is that the estimated entropy bounds the true entropy of the data from above. This is because among all distributions with a given mean and covariance, normal distribution has the largest entropy. Thus, the true coding length cannot be more than what the algorithm tries to minimize.
For any given segmentation of an image, this scheme yields the number of bits required to encode that image based on the given segmentation. Thus, among all possible segmentations of an image, the goal is to find the segmentation which produces the shortest coding length. This can be achieved by a simple agglomerative clustering method. The distortion in the lossy compression determines the coarseness of the segmentation and its optimal value may differ for each image. This parameter can be estimated heuristically from the contrast of textures in an image. For example, when the textures in an image are similar, such as in camouflage images, stronger sensitivity and thus lower quantization is required.
Histogram-based methods
Histogram-based methods are very efficient when compared to other image segmentation methods because they typically require only one pass through the pixels. In this technique, a histogram is computed from all of the pixels in the image, and the peaks and valleys in the histogram are used to locate the clusters in the image.[1] Color or intensity can be used as the measure.
A refinement of this technique is to recursively apply the histogram-seeking method to clusters in the image in order to divide them into smaller clusters. This is repeated with smaller and smaller clusters until no more clusters are formed.[1][5]
One disadvantage of the histogram-seeking method is that it may be difficult to identify significant peaks and valleys in the image. In this technique of image classification distance metric and integrated region matching are familiar.
Histogram-based approaches can also be quickly adapted to occur over multiple frames, while maintaining their single pass efficiency. The histogram can be done in multiple fashions when multiple frames are considered. The same approach that is taken with one frame can be applied to multiple, and after the results are merged, peaks and valleys that were previously difficult to identify are more likely to be distinguishable. The histogram can also be applied on a per pixel basis where the information result are used to determine the most frequent color for the pixel location. This approach segments based on active objects and a static environment, resulting in a different type of segmentation useful in Video tracking.
Edge detection
Edge detection is a well-developed field on its own within image processing. Region boundaries and edges are closely related, since there is often a sharp adjustment in intensity at the region boundaries. Edge detection techniques have therefore been used as the base of another segmentation technique.
The edges identified by edge detection are often disconnected. To segment an object from an image however, one needs closed region boundaries. The desired edges are the boundaries between such objects.
Segmentation methods can also be applied to edges obtained from edge detectors. Lindeberg and Li [6] developed an integrated method that segments edges into straight and curved edge segments for parts-based object recognition, based on a minimum description length (MDL) criterion that was optimized by a split-and-merge-like method with candidate breakpoints obtained from complementary junction cues to obtain more likely points at which to consider partitions into different segments.
Region-growing methods
The first region-growing method was the seeded region growing method. This method takes a set of seeds as input along with the image. The seeds mark each of the objects to be segmented. The regions are iteratively grown by comparing all unallocated neighbouring pixels to the regions. The difference between a pixel's intensity value and the region's mean, 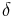 , is used as a measure of similarity. The pixel with the smallest difference measured this way is allocated to the respective region. This process continues until all pixels are allocated to a region.
, is used as a measure of similarity. The pixel with the smallest difference measured this way is allocated to the respective region. This process continues until all pixels are allocated to a region.
Seeded region growing requires seeds as additional input. The segmentation results are dependent on the choice of seeds. Noise in the image can cause the seeds to be poorly placed. Unseeded region growing is a modified algorithm that doesn't require explicit seeds. It starts off with a single region 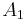 – the pixel chosen here does not significantly influence final segmentation. At each iteration it considers the neighbouring pixels in the same way as seeded region growing. It differs from seeded region growing in that if the minimum
– the pixel chosen here does not significantly influence final segmentation. At each iteration it considers the neighbouring pixels in the same way as seeded region growing. It differs from seeded region growing in that if the minimum 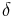 is less than a predefined threshold
is less than a predefined threshold 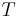 then it is added to the respective region
then it is added to the respective region 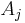 . If not, then the pixel is considered significantly different from all current regions
. If not, then the pixel is considered significantly different from all current regions 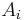 and a new region
and a new region 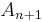 is created with this pixel.
is created with this pixel.
One variant of this technique, proposed by Haralick and Shapiro (1985),[1] is based on pixel intensities. The mean and scatter of the region and the intensity of the candidate pixel is used to compute a test statistic. If the test statistic is sufficiently small, the pixel is added to the region, and the region’s mean and scatter are recomputed. Otherwise, the pixel is rejected, and is used to form a new region.
A special region-growing method is called 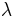 -connected segmentation (see also lambda-connectedness). It is based on pixel intensities and neighborhood-linking paths. A degree of connectivity (connectedness) will be calculated based on a path that is formed by pixels. For a certain value of
-connected segmentation (see also lambda-connectedness). It is based on pixel intensities and neighborhood-linking paths. A degree of connectivity (connectedness) will be calculated based on a path that is formed by pixels. For a certain value of 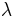 , two pixels are called
, two pixels are called 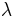 -connected if there is a path linking those two pixels and the connectedness of this path is at least
-connected if there is a path linking those two pixels and the connectedness of this path is at least 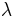 .
. 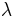 -connectedness is an equivalence relation.[7]
-connectedness is an equivalence relation.[7]
Split-and-merge methods
Split-and-merge segmentation is based on a quadtree partition of an image. It is sometimes called quadtree segmentation.
This method starts at the root of the tree that represents the whole image. If it is found non-uniform (not homogeneous), then it is split into four son-squares (the splitting process), and so on so forth. Conversely, if four son-squares are homogeneous, they can be merged as several connected components (the merging process). The node in the tree is a segmented node. This process continues recursively until no further splits or merges are possible.[8][9] When a special data structure is involved in the implementation of the algorithm of the method, its time complexity can reach 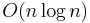 , an optimal algorithm of the method.[10]
, an optimal algorithm of the method.[10]
Partial differential equation-based methods
Using a partial differential equation (PDE)-based method and solving the PDE equation by a numerical scheme, one can segment the image. Curve propagation is a popular technique in this category, with numerous applications to object extraction, object tracking, stereo reconstruction, etc. The central idea is to evolve an initial curve towards the lowest potential of a cost function, where its definition reflects the task to be addressed. As for most inverse problems, the minimization of the cost functional is non-trivial and imposes certain smoothness constraints on the solution, which in the present case can be expressed as geometrical constraints on the evolving curve.
Parametric methods
Lagrangian techniques are based on parameterizing the contour according to some sampling strategy and then evolve each element according to image and internal terms. Such techniques are fast and efficient, however the original "purely parametric" formulation (due to Kass and Terzopoulos in 1987 and known as "snakes"), is generally criticized for its limitations regarding the choice of sampling strategy, the internal geometric properties of the curve, topology changes (curve splitting and merging), addressing problems in higher dimensions, etc.. Nowadays, efficient "discretized" formulations have been developed to address these limitations while maintaining high efficiency. In both cases, energy minimization is generally conducted using a steepest-gradient descent, whereby derivatives are computed using, e.g., finite differences.
Level set methods
The level set method was initially proposed to track moving interfaces by Osher and Sethian in 1988 and has spread across various imaging domains in the late nineties. It can be used to efficiently address the problem of curve/surface/etc. propagation in an implicit manner. The central idea is to represent the evolving contour using a signed function, where its zero level corresponds to the actual contour. Then, according to the motion equation of the contour, one can easily derive a similar flow for the implicit surface that when applied to the zero-level will reflect the propagation of the contour. The level set method encodes numerous advantages: it is implicit, parameter free, provides a direct way to estimate the geometric properties of the evolving structure, can change the topology and is intrinsic. Furthermore, they can be used to define an optimization framework as proposed by Zhao, Merriman and Osher in 1996. Therefore, one can conclude that it is a very convenient framework to address numerous applications of computer vision and medical image analysis.[11] Furthermore, research into various level set data structures has led to very efficient implementations of this method.
Graph partitioning methods
Graph partitioning methods can effectively be used for image segmentation. In these methods, the image is modeled as a weighted, undirected graph. Usually a pixel or a group of pixels are associated with nodes and edge weights define the (dis)similarity between the neighborhood pixels. The graph (image) is then partitioned according to a criterion designed to model "good" clusters. Each partition of the nodes (pixels) output from these algorithms are considered an object segment in the image. Some popular algorithms of this category are normalized cuts,[12] random walker,[13] minimum cut,[14] isoperimetric partitioning [15] and minimum spanning tree-based segmentation.[16]
Watershed transformation
The watershed transformation considers the gradient magnitude of an image as a topographic surface. Pixels having the highest gradient magnitude intensities (GMIs) correspond to watershed lines, which represent the region boundaries. Water placed on any pixel enclosed by a common watershed line flows downhill to a common local intensity minimum (LIM). Pixels draining to a common minimum form a catch basin, which represents a segment.
Model based segmentation
The central assumption of such an approach is that structures of interest/organs have a repetitive form of geometry. Therefore, one can seek for a probabilistic model towards explaining the variation of the shape of the organ and then when segmenting an image impose constraints using this model as prior. Such a task involves (i) registration of the training examples to a common pose, (ii) probabilistic representation of the variation of the registered samples, and (iii) statistical inference between the model and the image. State of the art methods in the literature for knowledge-based segmentation involve active shape and appearance models, active contours and deformable templates and level-set based methods.
Multi-scale segmentation
Image segmentations are computed at multiple scales in scale-space and sometimes propagated from coarse to fine scales; see scale-space segmentation.
Segmentation criteria can be arbitrarily complex and may take into account global as well as local criteria. A common requirement is that each region must be connected in some sense.
One-dimensional hierarchical signal segmentation
Witkin's seminal work[17][18] in scale space included the notion that a one-dimensional signal could be unambiguously segmented into regions, with one scale parameter controlling the scale of segmentation.
A key observation is that the zero-crossings of the second derivatives (minima and maxima of the first derivative or slope) of multi-scale-smoothed versions of a signal form a nesting tree, which defines hierarchical relations between segments at different scales. Specifically, slope extrema at coarse scales can be traced back to corresponding features at fine scales. When a slope maximum and slope minimum annihilate each other at a larger scale, the three segments that they separated merge into one segment, thus defining the hierarchy of segments.
Image segmentation and primal sketch
There have been numerous research works in this area, out of which a few have now reached a state where they can be applied either with interactive manual intervention (usually with application to medical imaging) or fully automatically. The following is a brief overview of some of the main research ideas that current approaches are based upon.
The nesting structure that Witkin described is, however, specific for one-dimensional signals and does not trivially transfer to higher-dimensional images. Nevertheless, this general idea has inspired several other authors to investigate coarse-to-fine schemes for image segmentation. Koenderink[19] proposed to study how iso-intensity contours evolve over scales and this approach was investigated in more detail by Lifshitz and Pizer.[20] Unfortunately, however, the intensity of image features changes over scales, which implies that it is hard to trace coarse-scale image features to finer scales using iso-intensity information.
Lindeberg[21][22] studied the problem of linking local extrema and saddle points over scales, and proposed an image representation called the scale-space primal sketch which makes explicit the relations between structures at different scales, and also makes explicit which image features are stable over large ranges of scale including locally appropriate scales for those. Bergholm proposed to detect edges at coarse scales in scale-space and then trace them back to finer scales with manual choice of both the coarse detection scale and the fine localization scale.
Gauch and Pizer[23] studied the complementary problem of ridges and valleys at multiple scales and developed a tool for interactive image segmentation based on multi-scale watersheds. The use of multi-scale watershed with application to the gradient map has also been investigated by Olsen and Nielsen[24] and been carried over to clinical use by Dam[25] Vincken et al.[26] proposed a hyperstack for defining probabilistic relations between image structures at different scales. The use of stable image structures over scales has been furthered by Ahuja[27][28] and his co-workers into a fully automated system. A fully automatic brain segmentation algorithm based on closely related ideas of multi-scale watersheds has been presented by Undeman and Lindeberg [29] and been extensively tested in brain databases.
These ideas for multi-scale image segmentation by linking image structures over scales have also been picked up by Florack and Kuijper.[30] Bijaoui and Rué[31] associate structures detected in scale-space above a minimum noise threshold into an object tree which spans multiple scales and corresponds to a kind of feature in the original signal. Extracted features are accurately reconstructed using an iterative conjugate gradient matrix method.
Semi-automatic segmentation
In this kind of segmentation, the user outlines the region of interest with the mouse clicks and algorithms are applied so that the path that best fits the edge of the image is shown.
Techniques like SIOX, Livewire, Intelligent Scissors or IT-SNAPS are used in this kind of segmentation.
Neural networks segmentation
Neural Network segmentation relies on processing small areas of an image using an artificial neural network[32] or a set of neural networks. After such processing the decision-making mechanism marks the areas of an image accordingly to the category recognized by the neural network. A type of network designed especially for this is the Kohonen map.
Pulse-coupled neural networks (PCNNs) are neural models proposed by modeling a cat’s visual cortex and developed for high-performance biomimetic image processing. In 1989, Eckhorn introduced a neural model to emulate the mechanism of cat’s visual cortex. The Eckhorn model provided a simple and effective tool for studying small mammal’s visual cortex, and was soon recognized as having significant application potential in image processing. In 1994, the Eckhorn model was adapted to be an image processing algorithm by Johnson, who termed this algorithm Pulse-Coupled Neural Network. Over the past decade, PCNNs have been utilized for a variety of image processing applications, including: image segmentation, feature generation, face extraction, motion detection, region growing, noise reduction, and so on. A PCNN is a two-dimensional neural network. Each neuron in the network corresponds to one pixel in an input image, receiving its corresponding pixel’s color information (e.g. intensity) as an external stimulus. Each neuron also connects with its neighboring neurons, receiving local stimuli from them. The external and local stimuli are combined in an internal activation system, which accumulates the stimuli until it exceeds a dynamic threshold, resulting in a pulse output. Through iterative computation, PCNN neurons produce temporal series of pulse outputs. The temporal series of pulse outputs contain information of input images and can be utilized for various image processing applications, such as image segmentation and feature generation. Compared with conventional image processing means, PCNNs have several significant merits, including robustness against noise, independence of geometric variations in input patterns, capability of bridging minor intensity variations in input patterns, etc.
Proprietary software
Several proprietary software packages are available for performing image segmentation
- Pac-n-Zoom Color has a proprietary software that color segments over 16 million colors at photographic quality. Manual available at http://www.accelerated-io.com/usr_man.htm [1]
- TurtleSeg is a free interactive 3D image segmentation tool. It supports many common medical image file formats and allows the user to export their segmentation as a binary mask or a 3D surface.
Several proprietary software packages are available for comparing the performance of segmentation methods with the state-of-the-art segmentation methods on standardized sets
- Prague On-line Texture Segmentation Benchmark[33]
- The Berkeley Segmentation Dataset and Benchmark[34]
Software
|
|||||||||||
Several open source software packages are available for performing image segmentation
- GIMP which includes among other tools SIOX (see Simple Interactive Object Extraction).
- ImageMagick segments using the Fuzzy C-Means algorithm.
- MITK has a program module for manual segmentation of gray-scale images.
- GRASS GIS has the program module i.smap for image segmentation.
- AForge.NET – an open source C# framework.
- Population open-source image processing library in C++
- Icy is an open-source image analysis software with several segmentation algorithms including an automatic "active contours" tool.
There are also software packages available free of charge for academic purposes:
See also
- Computer vision
- Data clustering
- Graph theory
- Histograms
- Image-based meshing
- K-means algorithm
- Pulse-coupled networks
- Range image segmentation
- Region growing
- Balanced histogram thresholding
External links
- Some sample code that performs basic segmentation, by Syed Zainudeen. University Technology of Malaysia.
References
- ^ a b c d e Linda G. Shapiro and George C. Stockman (2001): “Computer Vision”, pp 279-325, New Jersey, Prentice-Hall, ISBN 0-13-030796-3
- ^ Pham, Dzung L.; Xu, Chenyang; Prince, Jerry L. (2000). "Current Methods in Medical Image Segmentation". Annual Review of Biomedical Engineering 2: 315–337. doi:10.1146/annurev.bioeng.2.1.315. PMID 11701515.
- ^ Hossein Mobahi, Shankar Rao, Allen Yang, Shankar Sastry and Yi Ma. Segmentation of Natural Images by Texture and Boundary Compression, International Journal of Computer Vision (IJCV), 95 (1), pg. 86-98, Oct. 2011.
- ^ Shankar Rao, Hossein Mobahi, Allen Yang, Shankar Sastry and Yi Ma Natural Image Segmentation with Adaptive Texture and Boundary Encoding, Proceedings of the Asian Conference on Computer Vision (ACCV) 2009, H. Zha, R.-i. Taniguchi, and S. Maybank (Eds.), Part I, LNCS 5994, pp. 135--146, Springer.
- ^ Ohlander, Ron; Price, Keith; Reddy, D. Raj (1978). "Picture Segmentation Using a Recursive Region Splitting Method". Computer Graphics and Image Processing 8 (3): 313–333. doi:10.1016/0146-664X(78)90060-6.
- ^ T. Lindeberg and M.-X. Li "Segmentation and classification of edges using minimum description length approximation and complementary junction cues", Computer Vision and Image Understanding, vol. 67, no. 1, pp. 88--98, 1997.
- ^ L. Chen, H.D. Cheng, and J. Zhang, Fuzzy subfiber and its application to seismic lithology classification, Information Sciences: Applications, Vol 1, No 2, pp 77-95, 1994.
- ^ S.L. Horowitz and T. Pavlidis, Picture Segmentation by a Directed Split and Merge Procedure, Proc. ICPR, 1974, Denmark, pp.424-433.
- ^ S.L. Horowitz and T. Pavlidis, Picture Segmentation by a Tree Traversal Algorithm, Journal of the ACM, 23 (1976), pp. 368-388.
- ^ L. Chen, The lambda-connected segmentation and the optimal algorithm for split-and-merge segmentation, Chinese J. Computers, 14(1991), pp 321-331
- ^ S. Osher and N. Paragios. Geometric Level Set Methods in Imaging Vision and Graphics, Springer Verlag, ISBN 0387954880, 2003.
- ^ Jianbo Shi and Jitendra Malik (2000): "Normalized Cuts and Image Segmentation", IEEE Transactions on pattern analysis and machine intelligence, pp 888-905, Vol. 22, No. 8
- ^ Leo Grady (2006): "Random Walks for Image Segmentation", IEEE Transactions on Pattern Analysis and Machine Intelligence, pp. 1768–1783, Vol. 28, No. 11
- ^ Z. Wu and R. Leahy (1993): "An optimal graph theoretic approach to data clustering: Theory and its application to image segmentation", IEEE Transactions on Pattern Analysis and Machine Intelligence, pp. 1101–1113, Vol. 15, No. 11
- ^ Leo Grady and Eric L. Schwartz (2006): "Isoperimetric Graph Partitioning for Image Segmentation", IEEE Transactions on Pattern Analysis and Machine Intelligence, pp. 469–475, Vol. 28, No. 3
- ^ C. T. Zahn (1971): "Graph-theoretical methods for detecting and describing gestalt clusters", IEEE Transactions on Computers, pp. 68–86, Vol. 20, No. 1
- ^ Witkin, A. P. "Scale-space filtering", Proc. 8th Int. Joint Conf. Art. Intell., Karlsruhe, Germany,1019–1022, 1983.
- ^ A. Witkin, "Scale-space filtering: A new approach to multi-scale description," in Proc. IEEE Int. Conf. Acoust., Speech, Signal Processing (ICASSP), vol. 9, San Diego, CA, Mar. 1984, pp. 150–153.
- ^ Koenderink, Jan "The structure of images", Biological Cybernetics, 50:363–370, 1984
- ^ Lifshitz, L. and Pizer, S.: A multiresolution hierarchical approach to image segmentation based on intensity extrema, IEEE Transactions on Pattern Analysis and Machine Intelligence, 12:6, 529–540, 1990.
- ^ Lindeberg, T.: Detecting salient blob-like image structures and their scales with a scale-space primal sketch: A method for focus-of-attention, International Journal of Computer Vision, 11(3), 283–318, 1993.
- ^ Lindeberg, Tony, Scale-Space Theory in Computer Vision, Kluwer Academic Publishers, 1994, ISBN 0-7923-9418-6
- ^ Gauch, J. and Pizer, S.: Multiresolution analysis of ridges and valleys in grey-scale images, IEEE Transactions on Pattern Analysis and Machine Intelligence, 15:6 (June 1993), pages: 635–646, 1993.
- ^ Olsen, O. and Nielsen, M.: Multi-scale gradient magnitude watershed segmentation, Proc. of ICIAP 97, Florence, Italy, Lecture Notes in Computer Science, pages 6–13. Springer Verlag, September 1997.
- ^ Dam, E., Johansen, P., Olsen, O. Thomsen,, A. Darvann, T. , Dobrzenieck, A., Hermann, N., Kitai, N., Kreiborg, S., Larsen, P., Nielsen, M.: "Interactive multi-scale segmentation in clinical use" in European Congress of Radiology 2000.
- ^ Vincken, K., Koster, A. and Viergever, M.: Probabilistic multiscale image segmentation, IEEE Transactions on Pattern Analysis and Machine Intelligence, 19:2, pp. 109–120, 1997.]
- ^ M. Tabb and N. Ahuja, Unsupervised multiscale image segmentation by integrated edge and region detection, IEEE Transactions on Image Processing, Vol. 6, No. 5, 642–655, 1997.
- ^ E. Akbas and N. Ahuja, "From ramp discontinuities to segmentation tree"
- ^ C. Undeman and T. Lindeberg (2003) "Fully Automatic Segmentation of MRI Brain Images using Probabilistic Anisotropic Diffusion and Multi-Scale Watersheds", Proc. Scale-Space'03, Isle of Skye, Scotland, Springer Lecture Notes in Computer Science, volume 2695, pages 641--656.
- ^ Florack, L. and Kuijper, A.: The topological structure of scale-space images, Journal of Mathematical Imaging and Vision, 12:1, 65–79, 2000.
- ^ Bijaoui, A., Rué, F.: 1995, A Multiscale Vision Model, Signal Processing 46, 345
- ^ Mahinda Pathegama & Ö Göl (2004): "Edge-end pixel extraction for edge-based image segmentation", Transactions on Engineering, Computing and Technology, vol. 2, pp 213–216, ISSN 1305-5313
- ^ Haindl, M. – Mikeš, S. Texture Segmentation Benchmark, Proc. of the 19th Int. Conference on Pattern Recognition. IEEE Computer Society, 2008, pp. 1–4 ISBN 978-1-4244-2174-9 ISSN 1051-4651
- ^ D.Martin; C. Fowlkes and D. Tal and J. Malik (July 2001). "A Database of Human Segmented Natural Images and its Application to Evaluating Segmentation Algorithms and Measuring Ecological Statistics". Proc. 8th Int'l Conf. Computer Vision. 2. pp. 416–423.
- Notes
- 3D Entropy Based Image Segmentation
- Frucci, Maria; Sanniti di Baja, Gabriella (2008). "From Segmentation to Binarization of Gray-level Images". Journal of Pattern Recognition Research 3 (1): 1–13. http://www.jprr.org/index.php/jprr/article/view/54/16.